3D images allow detailed insight into grasses
Tao Ju and Dan Zeng, a graduate student in Ju's lab, demonstrate how to measure a wide array of sorghum panicle traits, some of which had never been quantified before

In a collaboration with scientists at the Donald Danforth Plant Science Center, Tao Ju and Dan Zeng, both in the Department of Computer Science & Engineering in the McKelvey School of Engineering at Washington University in St. Louis, have developed an algorithm that analyzes the shape of a sorghum panicle from X-ray computed tomography.
Their work, which was published in and on the cover of New Phytologist in May, demonstrates how to measure a wide array of sorghum panicle traits, some of which had never been quantified before.
Scientists want to study the characteristics of the panicle to learn more about the plant's yield, nutritional quality and disease resistance. Until now, they had to study the panicle manually using rulers, scales and 2D images, which limited accuracy. The 3D images provide precise quantitative data of the panicle that will allow scientists to determine differences between each of the sorghum varieties.
Zeng, a doctoral student in Ju's lab, designed the algorithm to process dozens of 3D images taken by Danforth Center scientists of the sorghum panicle, a foot-long, flowering structure containing the plant's tightly-packed seeds. Two-dimensional X-ray scans are taken from various angles with the panicle on a turntable, to get the architectural traits from the image using computerized tomography (CT).
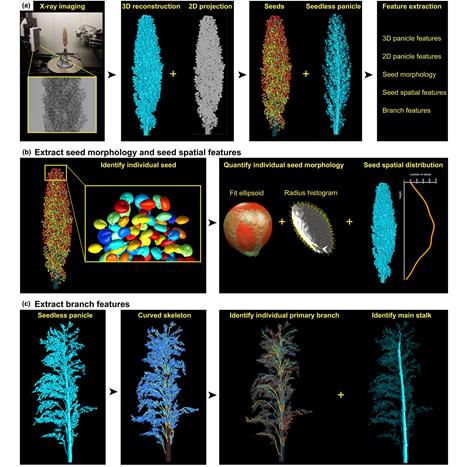
Prior to Zeng's tool, to measure some traits of the panicle, such as the number of seeds and branches, one would need to physically take apart the panicle one seed or branch at a time. This manual approach is not only labor-intensive, but it also destroys the structure of the panicle and makes further evaluation difficult.
Zeng's algorithm analyzes the branches of the panicle from the CT image.
"The algorithm extracts branches from the image as 3D curves, and then traces along the curves to count the branches and measure their geometric properties, such as length and curvature," Zeng said. "This gets the traits directly from the image of the plant, is less time-consuming, and doesn't alter the geometry of the panicle."
"With these measurements, we get a more refined characterization of the panicle, which allows us to build a better correlation between plant shape and genetics," said Ju, also vice dean for research. "Perhaps more importantly, the imaging pipeline and computational tools developed from this study get us one step closer towards high-throughput, high-resolution characterization of complex plant forms, which would enable biologists and agricultural researchers to answer more questions that were difficult to answer previously without 3D imaging."
Read the full release from the Donald Danforth Plant Science Center here.
Li M, Shao M‐R, Zeng D, Ju T, Kellogg EA and Topp CN. Comprehensive 3D phenotyping reveals continuous morphological variation across genetically diverse sorghum inflorescences. New Phytol, 226: 1873-1885. doi:10.1111/nph.16533