New microscopy method provides unprecedented look at amyloid protein structure
Measuring location, orientation of single molecules could give new insights into Alzheimer’s, Parkinson’s
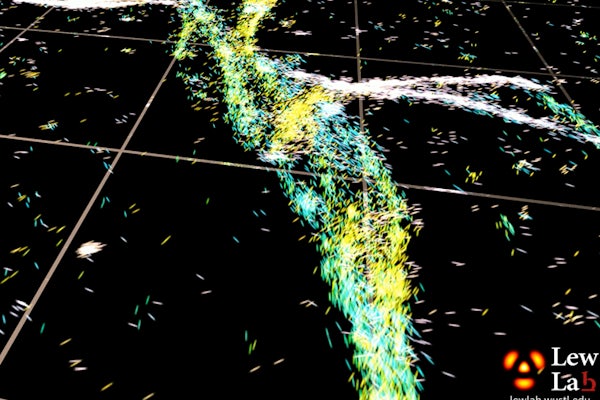
Neurodegenerative diseases such as Alzheimer’s and Parkinson’s are often accompanied by amyloid proteins in the brain that have become clumped or misfolded. At Washington University in St. Louis, a newly developed technique that measures the orientation of single molecules is enabling, for the first time, optical microscopy to reveal nanoscale details about the structures of these problematic proteins.
Research from the lab of Matthew Lew, assistant professor in the Preston M. Green Department of Electrical & Systems Engineering at the McKelvey School of Engineering, describing this new approach was published in Optica, The Optical Society’s journal for high impact research.
“Neurodegenerative disorders such as Alzheimer’s and Parkinson’s diseases are leading causes of death all over the world,” said Tianben Ding, a PhD student in Lew’s lab and a co-author of the new paper. “We hope our single-molecule orientation imaging approach can provide new insights into amyloid structure and possibly contribute future development of effective therapeutics against the diseases.”
Biological and chemical processes are driven by complicated movements and interactions between molecules. Although most amyloid proteins may be non-toxic, the misfolding of even a few could eventually kill many neurons.
“We need imaging technologies that can watch these molecular movements in living systems to understand the fundamental biological mechanisms of disease,” Lew said. “Amyloid and prion-type diseases like Alzheimer’s, Parkinson’s and diabetes are our first targets for this technology, but we see it being applied in many other areas too.”
Selecting the best microscope
Lew’s lab has developed several microscopy methods that measure the orientation and location of fluorescent molecules attached to single proteins. The orientation information is obtained by measuring not only the location of fluorescence in the sample but also characteristics of that light, such as polarization, which are typically ignored in most other microscopy approaches.
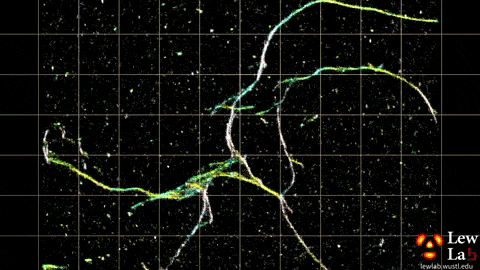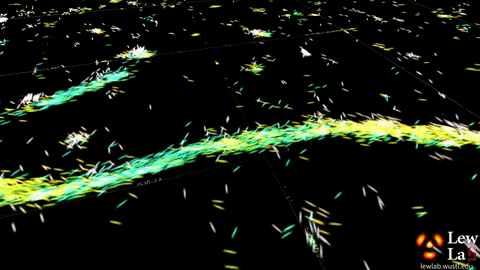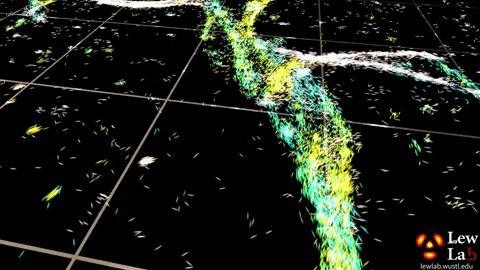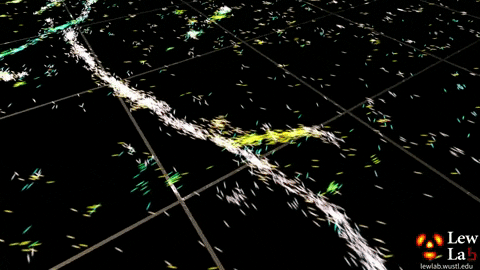
“By measuring the orientations of single molecules bound to amyloid aggregates, this microscope enabled us to map differences in amyloid structure organization that cannot be detected by standard localization microscopes,” said Tingting Wu, a co-author of the paper.
The researchers used the powerful microscopy approach to measure the orientations of single fluorescent molecules bound to amyloid aggregates. Because there is no artificial linker between the fluorescent probes and amyloid surfaces, the probes’ binding orientation to the amyloid surfaces conveys information about how the amyloid protein itself is organized.
The researchers quantified how the orientations of each fluorescent molecule varied each time one attached to an amyloid protein. Differences in these binding behaviors can be attributed to structure differences between amyloid aggregates. Because the method provides single-molecule information, the researchers could observe nanoscale differences between amyloid structures without averaging out details of local features.
Opportunities for long-term studies
“We plan to extend the method to monitor nanoscale changes within and between amyloid structures as they organize over hours to days,” said Ding. “Long-term studies of amyloid aggregates may reveal new correlations between how amyloid proteins are organized and how quickly they grow or spontaneously dissolve.”
The researchers note that the set-up they used for orientation-localization microscopy consisted of commercially available parts that are accessible to anyone performing single-molecule super-resolution microscopy. Their analysis code is available online at: https://github.com/Lew-Lab/RoSE-O.
“In optical microscopy and imaging, scientists and engineers have been pushing the boundaries of imaging to be faster, probe deeper and have higher resolution,” Lew said. “Our work shows that one can shed light on fundamental processes in biology by, instead, focusing on molecular orientation, which can reveal details about the inner workings of biology that cannot be visualized by traditional microscopy.”
Read the full release on the OSA website.
T. Ding, T. Wu, H. Mazidi, O. Zhang, M. Lew. Single-molecule orientation localization microscopy for resolving structural heterogeneities between amyloid fibrils. Optica, June 4, 2020. DOI: doi.org/10.1364/OPTICA.388157.




