Personalized medicine for lung cancer gets closer look
Abhinav Jha’s latest NIH grant another step toward early prediction of therapy response in patients with lung cancer

Lung cancer is the leading cause of mortality among all cancers in the United States. About 85% of newly diagnosed lung-cancer cases are non-small cell lung cancer (NSCLC), of which up to half are stage 3 at time of diagnosis. Long-term outcomes are poor at this stage, with only 5%-10% of the patients with stage 3B surviving five years after diagnosis.
At the same time, there are multiple options to treat this disease, including chemotherapy, radiation and surgery.
“While there are a lot of treatment options available, there is also a lot of heterogeneity in this patient population, and it’s not clear which treatment is best for each patient,” said Abhinav Jha, assistant professor of biomedical engineering in the McKelvey School of Engineering at Washington University in St. Louis.
“Thus, there’s a need for personalized treatment strategies,” he continued. “Imaging provides a mechanism to personalize treatment by probing if there are changes occurring in the tumor in response to treatment. By personalizing the treatment, we can reduce side effects and monetary costs, improve quality of life and potentially improve clinical outcomes for the patient.”
Reliable tumor segmentation is key to quantifying changes occurring within tumors. Jha recently received a $493,000 grant from the National Institutes of Health (NIH) to continue investigating ways to automatically identify and segment NSCLC tumors imaged by the widely used positron enhanced tomography (PET) technique—an important first step to someday predicting the effectiveness of therapy in patients with NSCLC.
“Our idea is to extract quantitative features from images of the tumor regions and see if [the tumor] is changing in response to treatment,” said Jha, noting that PET is well-suited for the task because it probes tumors at the molecular level, so any changes during therapy can be observed early.
However, segmenting tumors in PET images is not without its challenges, Jha said. For example, PET scans have limited resolution, making it difficult to accurately delineate tumor boundaries.
Jha’s physics-guided deep learning-based method overcomes these challenges by using simulations to generate thousands of PET images where the tumor boundaries are known. He can then use these images to train a deep-learning algorithm for segmenting tumors. The algorithm is then fine-tuned on a small amount of clinical data.
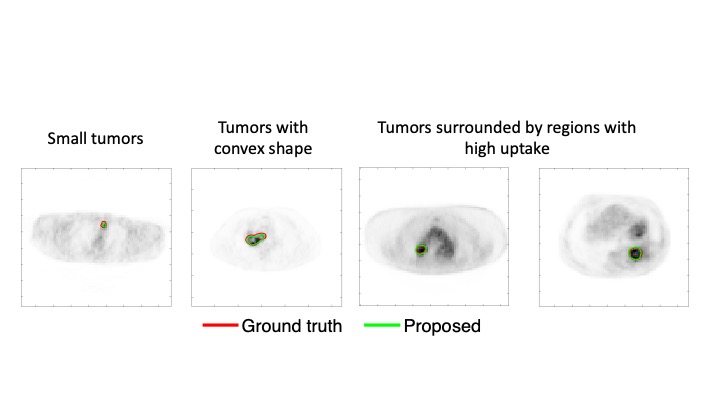
In a recently published paper, Jha showed that the method is able to accurately segment tumors, while requiring low amounts of clinical data for training, and is observed to generalize across different scanners.
Jha and his collaborators continued to advance in this area, and recently also proposed another approach that addressed the issue of tissue-fraction effects in PET segmentation, namely that voxels in a PET image can be part tumor and part background. They have demonstrated that this new method was able to accurately segment tumors from PET images acquired as part of a multicenter American College of Radiology Imaging Network (ACRIN) trial. Further, their method significantly outperformed widely used conventional PET segmentation methods.
Jha’s latest NIH grant is part of the National Institute of Biomedical Imaging and Bioengineering (NIBIB) R56 program for short-term, high-priority projects. At the end of this one-year grant, Jha aims to advance his segmentation framework from a 2D to a 3D convolutional neural network segmentation technique that can automatically locate and segment NSCLC tumors. A 3D approach is more useful because it can delineate the entire tumor in one pass rather than having to delineate it slice-by-slice as done in a 2D system.
In addition, the tumor segmentation method will be validated using a combination of realistic simulations and actual patient images obtained from ACRIN clinical trials.
Jha has assembled an impressive multi-disciplinary team for this project, including fellow faculty from the WashU School of Medicine, who will assist with clinical aspects of the study. Co-investigators on the grant include Barry Siegel, MD, professor of radiology and medicine and former chief of the Division of Nuclear Medicine; Ramaswamy Govindan, MD, Anheuser Busch Endowed Chair in Medical Oncology; Joyce Mhlanga, MD, assistant professor of radiology; Tyler Fraum, MD, assistant professor of radiology; and Richard Laforest, professor of radiology.



