Statistical theory, deep learning, lead to novel approach to differentiate tumors
Jha lab develops Bayesian approach to segment tumors in PET scans
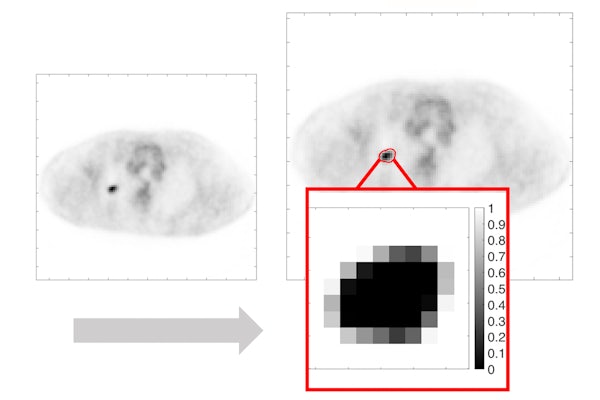
PET scans are frequently conducted in patients with cancer for clinical tasks. However, differentiating the tumor from healthy tissue in these images is challenging.
A biomedical engineer at Washington University in St. Louis has applied statistical theory and deep learning to differentiate a tumor from normal tissue in PET images. The method outperformed existing methods and more accurately differentiated tumors from normal tissue even when the tumor was small, as evaluated using realistic simulations and using clinical images from patients with stage 2-B/3 non-small cell lung cancer from a multi-center clinical trial. Results of the work are published in the special issue on early-career researchers in Physics in Medicine & Biology May 17, 2021.
The work was a team effort by Abhinav Jha, assistant professor of biomedical engineering in the McKelvey School of Engineering; Ziping Liu, a doctoral student in his lab; along with Joyce C. Mhlanga, MD, assistant professor of radiology; Paul-Robert Derenoncourt, MD, a resident physician in radiology; Richard Laforest, professor of radiology; and Barry A. Siegel, MD, professor of radiology and former chief of the Division of Nuclear Medicine, all at Washington University School of Medicine.
The key premise behind this work is that while tumors are continuous regions, PET images are discrete and consist of voxels, a discretized unit in the 3D space. Thus, a single voxel may contain both tumor and normal tissue. Most conventional methods classify a voxel as either tumor or normal tissue and are inherently limited in accounting for this issue. To address this, the team developed a Bayesian approach, a statistical model that estimated the fractional tumor volume within each voxel.
“The key idea is that we don’t just learn if a voxel belongs to the tumor or not,” Jha said. “The voxel can be part tumor and part normal. The novelty is that we can estimate how much of the voxel is the tumor.
“We asked the question that given a population of patient images, what would be the estimator that would minimize a certain measure of error between the true and estimated tumor fraction volumes?” Jha said. “The answer led to an estimator referred to as posterior mean. To derive this estimator, we used a deep-learning approach.”
The method was evaluated, first using realistic simulation studies, and then using data from the ACRIN 6668, a multicenter clinical trial.
“We gave the algorithm a population of images then told the algorithm that this much of the pixel is tumor and this much is not,” Jha said. “After that, we trained the network to estimate the tumor fraction volume. Once it learned how to do it, we tested the method with independent data. The method yielded accurate performance in both clinical and simulated images.”
Jha and his team plan to use this algorithm to improve quantitative PET.
“We want to look at the extracting features, such as volume of the images, and see if they can help us to make better decisions about patient care,” he said.
Another application is tumor delineation for radiation therapy planning.
“It’s a quality-of-life issue for the patient, and helping to answer those questions would be satisfying and rewarding.”
Liu Z, Mhlanga JC, Laforest R, Derenoncourt P-R, Siegel BA, Jha AK. A Bayesian approach to tissue-fraction estimation for oncological PET segmentation. Physics in Medicine & Biology, Accepted Manuscript online May 17, 2021. https://iopscience.iop.org/article/10.1088/1361-6560/ac01f4/meta
Funding for this work was provided by the National Institute of Biomedical Imaging and Bioengineering Trailblazer R21 Award (R21-EB024647), R01 EB031051 and the NVIDIA GPU grant. Computational resources were provided by the Washington University Center for High-Performance Computing.





